render¶
Opening Files¶
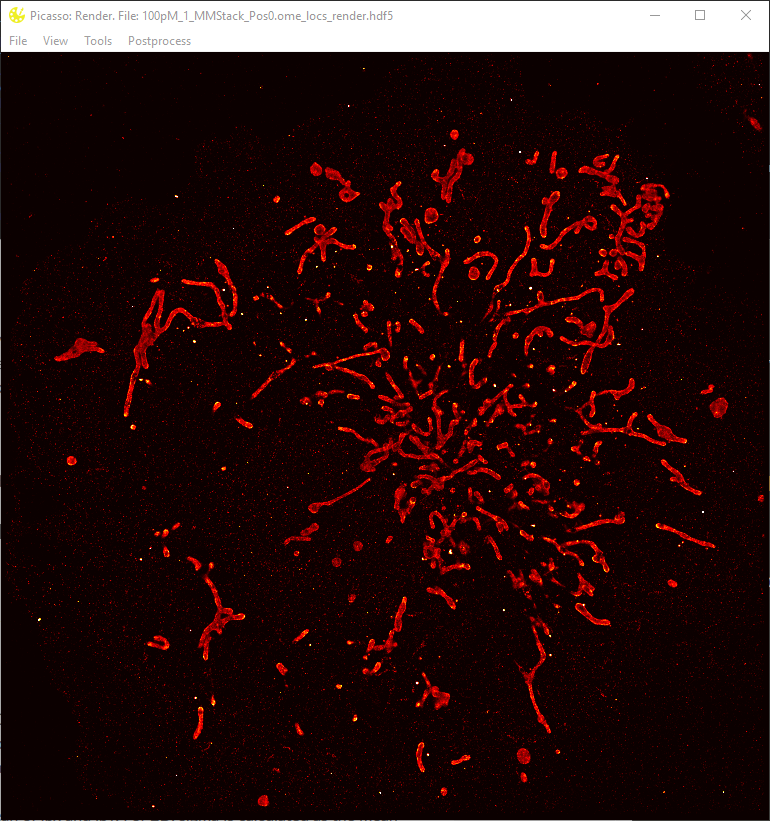
- Rendering of the super-resolution image: In
Picasso: Render, open a movie file by dragging a localization file (ending with ‘.hdf5’) into the window or by selectingFile > Open. The super-resolution image will be rendered automatically. A region of choice can be zoomed into by a rectangular selection using the left mouse button. The ‘View’ menu contains more options for zooming and panning. - (Optional) Adjust rendering options by selecting
View > Display Settings. The field ‘Oversampling’ defines the number of super-resolution pixels per camera pixel. The contrast settingsMin. DensityandMax. Densitydefine at which number of localizations per super-resolution pixel the minimum and maximum color of the colormap should be applied. - (Optional) For multiplexed image acquisition, open HDF5 localization files from other channels subsequently. Alternatively, drag and drop all HDF5 files to be displayed simultaneously.
Drift Correction¶
Picasso offers two procedures to correct for drift: an RCC algorithm (option A), and use of specific structures in the image as drift markers (option B). Although option A does not require any additional sample preparation, option B depends on the presence of either fiducial markers or inherently clustered structures in the image. On the other hand, option B often supports more precise drift estimation and thus allows for higher image resolution. To achieve the highest possible resolution (ultra-resolution), we recommend consecutive applications of option A and multiple rounds of option B. The drift markers for option B can be features of the image itself (e.g., protein complexes or DNA origami) or intentionally included markers (e.g., DNA origami or gold nanoparticles). When using DNA origami as drift markers, the correction is typically applied in two rounds: first, with whole DNA origami structures as markers, and, second, using single DNA-PAINT binding sites as markers. In both cases, the precision of drift correction strongly depends on the number of selected drift markers.
Redundant cross-correlation drift correction¶
- In
Picasso: Render, selectPostprocess > Undrift by RCC. - A dialog will appear asking for the segmentation parameter. Although the default value, 1,000 frames, is a sensible choice for most movies, it might be necessary to adjust the segmentation parameter of the algorithm, depending on the total number of frames in the movie and the number of localizations per frame. A smaller segment size results in better temporal drift resolution but requires a movie with more localizations per frame.
- After the algorithm finishes, the estimated drift will be displayed in a pop-up window, and the display will show the drift-corrected image.
Marker-based drift correction¶
- In
Picasso: Render, pick drift markers as described in Picking of regions of interest. Use thePick similaroption to automatically detect a large number of drift markers similar to a few manually selected ones. - If the structures used as drift markers have an intrinsic size larger than the precision of individual localizations (e.g., DNA origami, large protein complexes), it is critical to select a large number of structures. Otherwise, the statistic for calculating the drift in each frame (the mean displacement of localization to the structure’s center of mass) is not valid.
- Select
Postprocess > Undrift from pickedto compute and apply the drift correction. - (Optional) Save the drift-corrected localizations by selecting
File > Save localizations.
Picking of regions of interest¶
- Manual selection. Open
Picasso: Renderand load the localization HDF5 file to be processed. - Switch the active tool by selecting
Tools > Pick. The mouse cursor will now change to a circle. Alternatively, openTools > Tools Settingsto change the shape into a rectangle. - Set the size of the pick circle by adjusting the
Diameterfield in the tool settings dialog (Tools > Tools Settings). Alternatively, chooseWidthfor a rectangular shape. - Pick regions of interest using the circular mouse cursor by clicking the left mouse button. All localizations within the circle will be selected for further processing.
- (Optional) Automated region of interest selection. Select
Tools > Pick similarto automatically detect and pick structures that have similar numbers of localizations and RMS deviation (RMSD) from their center of mass than already-picked structures. The upper and lower thresholds for these similarity measures are the respective standard deviations of already-picked regions, scaled by a tunable factor. This factor can be adjusted using the fieldTools > Tools Settings > Pick similar ± range. To display the mean and standard deviation of localization number and RMSD for currently picked regions, selectView > Show infoand clickCalculate info below. - (Optional) Exporting of pick information. All localizations in picked regions can be saved by selecting
File > Save picked localizations. The resulting HDF5 file will contain a new integer columngroupindicating to which pick each localization is assigned. - (Optional) Statistics about each pick region can be saved by selecting
File > Save pick properties. The resulting HDF5 file is not a localization file. Instead, it holds a data set calledgroupsin which the rows show statistical values for each pick region. - (Optional) The picked positions and diameter itself can be saved by selecting
File > Save pick regions. Such saved pick information can also be loaded intoPicasso: Renderby selectingFile > Load pick regions.
3D rotation window¶
The 3D rotation window allows the user to render 3D localization data. To use it, select a single pick region (Tools > Pick) and click View > Update rotation window. Some of the display settings (colors, blur method, etc.) are automatically uploaded to the rotation window.
The user may perform multiple actions in the rotation window, including: saving rotated localizations, building animations (.mp4 format), rotating by a specified angle, etc.
Rotation around z-axis is available by pressing Ctrl/Command. Rotation axis can be frozen by pressing x/y/z to freeze around the corresponding axes (to freeze around the z-axis, Ctrl/Command must be pressed as well).
There are several things to keep in mind when using the rotation window. Firstly, using individual localization precision is very slow and is not recommended as a default blur method. Also, the size of the rotation window can be altered, however, if it becomes too large, rendering may start to lag.
RESI¶
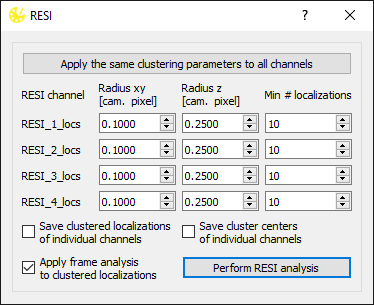
In Picasso 0.6.0, a new RESI (Resolution Enhancement by Sequential Imaging) dialog was introduced. It allows for a substantial resolution boost by sequential imaging of a single target with multiple labels with Exchange-PAINT (Reinhardt, et al., Nature, 2023. DOI: 10.1038/s41586-023-05925-9).
To use RESI, prepare your individual RESI channels (localization, undrifting, filtering and alignment). Load such localization lists into Picasso Render and open Postprocess > RESI. The dialog shown above will appear. Each channel will be clustered using the SMLM clusterer (other clustering algorithms could be applied as well although only the SMLM clusterer is implemented for RESI in Picasso). Clustering parameters can be defined for each RESI channel individually, although it is possible to apply the same parameters to all channels by clicking Apply the same clustering parameters to all channels, which will copy the clustering parameters from the first row and paste it to all other channels.
Next, the user needs to specify whether or not to save clustered localizations or cluster centers from each of the RESI channels individually, and whether to apply basic frame analysis (to minimize the effect of sticking events). For the explanation of the parameters, see SMLM clusterer below.
Upon clicking Perform RESI analysis, each of the loaded channels is clustered, cluster centers are extracted and combined from all RESI channels to create the final RESI file.
Dialogs¶
Display Settings¶
Allows to change the display settings. Open via View > Display Settings.
General¶
Adjust the general display settings.
Zoom¶
Set the magnification factor.
Oversampling¶
Set the oversampling. Choose dynamic to automatically adjust to current window size when zooming.
Minimap¶
Click show minimap to display a minimap in the upper left corner to localize where the current field of view is within the image.
Contrast¶
Define the minimum and maximum density of the and select a colormap. Over 100 colormaps are available. The last option Custom requires the user to load their own .npy file containg a numpy array with a custom colormap. The selected colormap will be saved when closing render.
Blur¶
Select a blur method. Available options are: * None * One-Pixel-Blur * Individual Localization Precision * Individual Localization Precision, iso
Camera¶
Select the pixel size of the camera. This will be automatically set to a default value or the value specified in the *.yaml file.
Scale Bar¶
Activate scalebar. The length of the scale bar is calculated with the Pixel Size set in the Camera dialog. Activate Print scale bar length to additionally print the length.
Render properties¶
This allows rendering properties by color.
Show Info¶
Displays the info dialog.
Display¶
Shows the image width/height, the coordinates, and dimensions of the current FoV.
Movie¶
Displays the median fit precision of the dataset. Clicking on Calculate allows calculating the precision via the NeNA approach. See DOI: 10.1007/s00418-014-1192-3.
Field of view¶
Shows the number of localizations in the current FoV.
Picks¶
Allows calculating statistics about the picked localizations. Press Calculate info below to calculate. Ignore dark times allows treating consecutive localizations as on, even if there are localizations (specified by the parameter) missing between them. When defining the number of units per pick, you can calibrate the influx rate via Calibrate influx. A histogram of the dark and bright time can be plotted when clicking Histograms.